Note that if you want to save your credits, remember to Terminate to stop
and end the runs.
What you will do
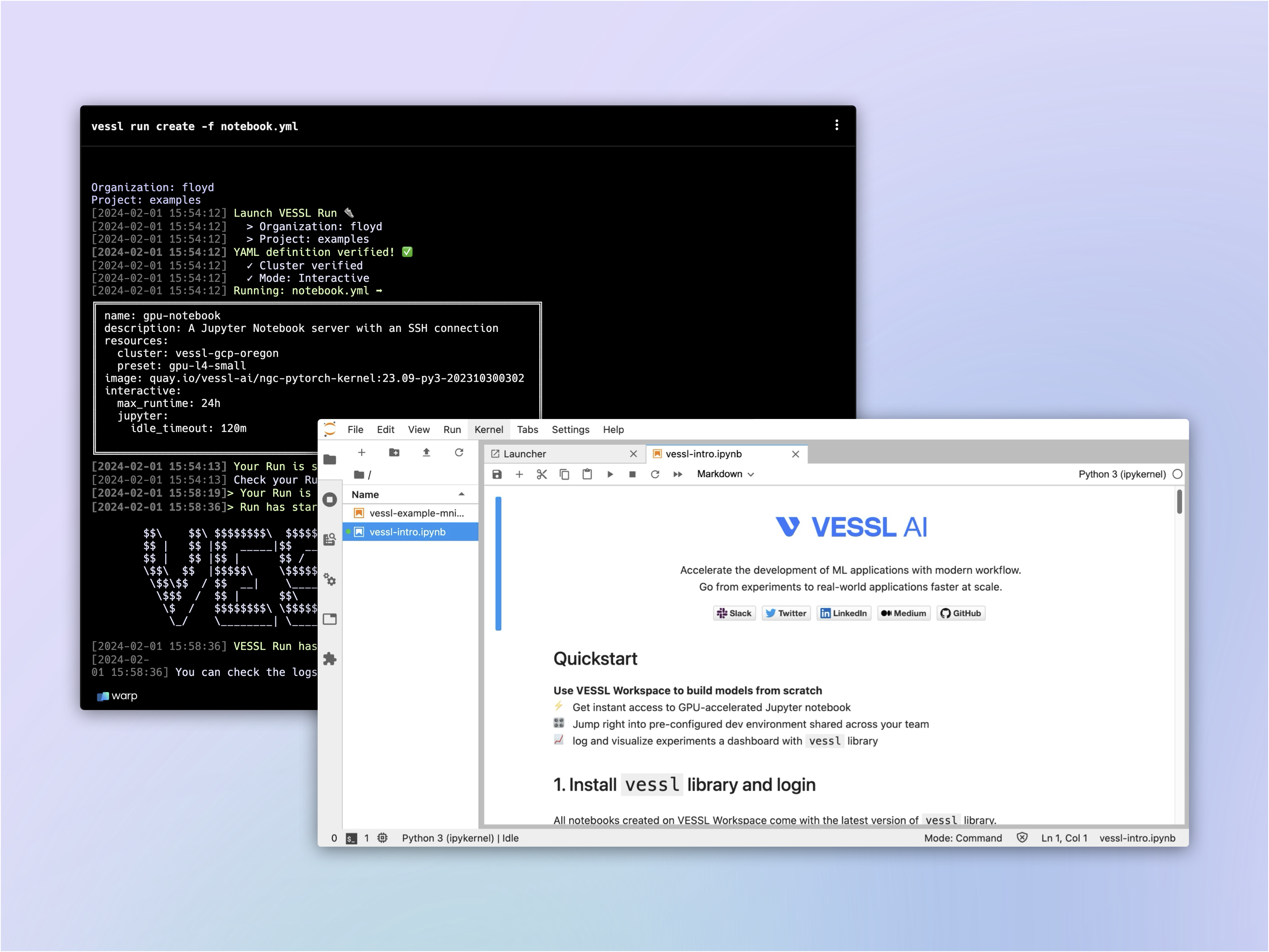
- Launch a GPU-accelerated interactive workload
- Set up a Jupyter Notebook
- Use SSH to connect to the workload
Writing the YAML file
Let’s start by creatingnotebook.yaml file and fill it in.
Spin up a workload
Let’s repeat the steps from Quickstart and define the compute resource and runtime environment for our workload. Again, we will use the L4 instance from our managed cloud and the latest PyTorch container from NVIDIA NGC.
Configure an interactive run
By default, workloads launched with VESSL Run are batch jobs like the one we launched in our Quickstart example. On the other hand, interactive runs are essentially virtual machines running on GPUs for live interaction with your models and datasets.You can enable this with the
interactive key, followed by the jupyter key. Interactive runs come with a default field for idle culler which automatically shuts down user notebook servers when they have not been used for a certain period.max_runtime works with idle_timeout as an additional measure to prevent resource overuseRunning the workload
Now that we have a completed YAML, we can once again run the workload withvessl run.
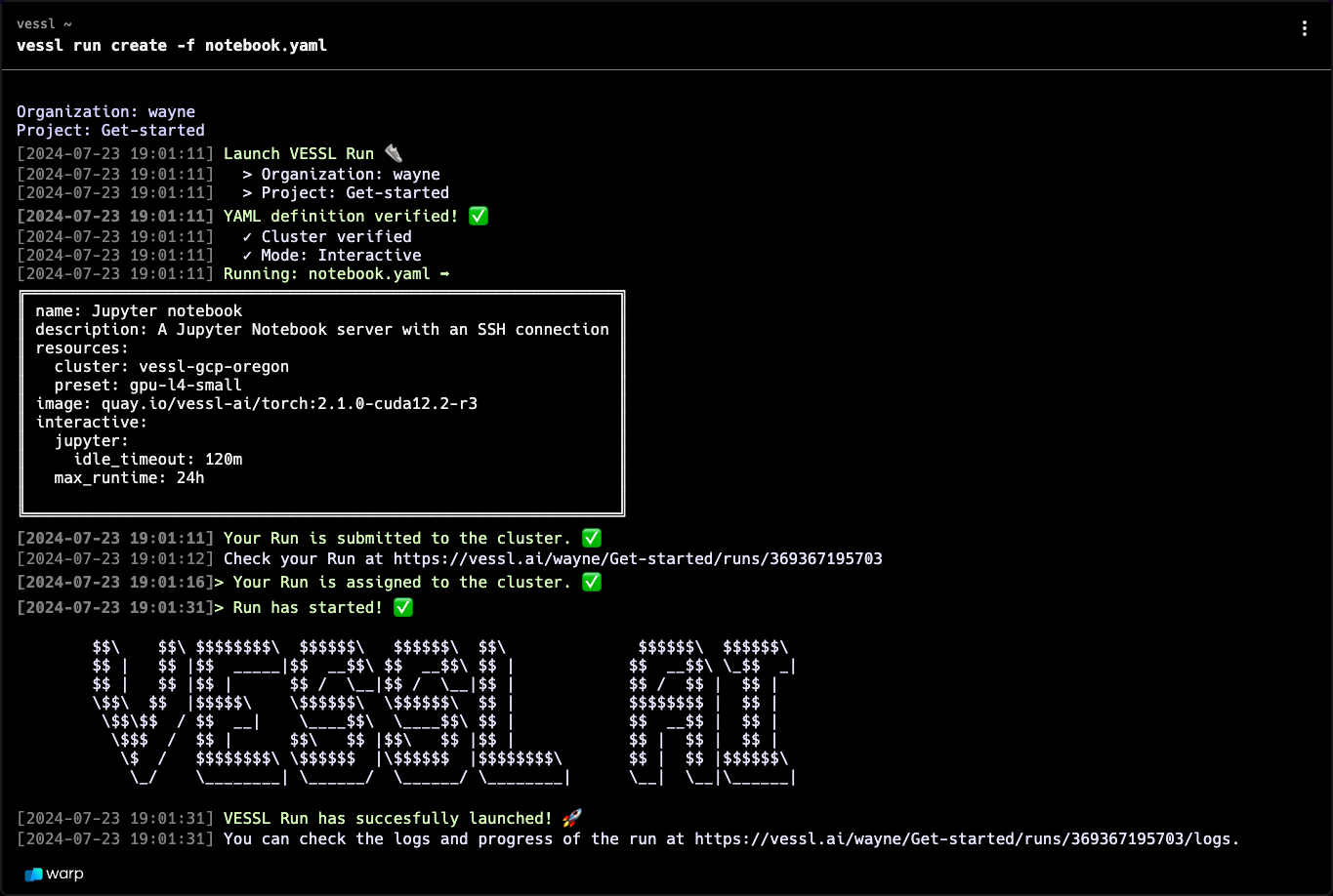
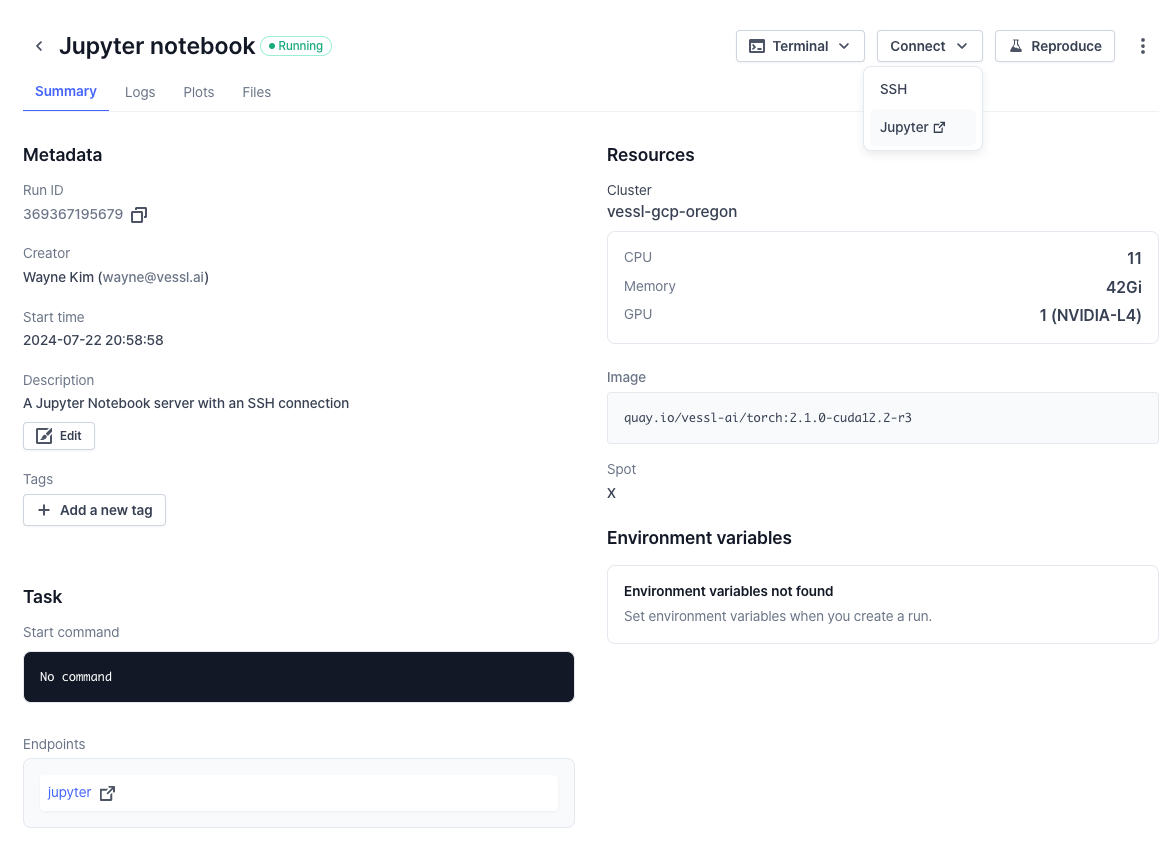
Create an SSH connection
An interactive run is essentially a GPU-accelerated workload on a cloud with a port and an endpoint for live interactions. This means you can access the remote workload using SSH.Connect via SSH
Use the workload address from the Run Summary page to connect. You are ready
to use VS Code or an IDE
of your choice for remote development.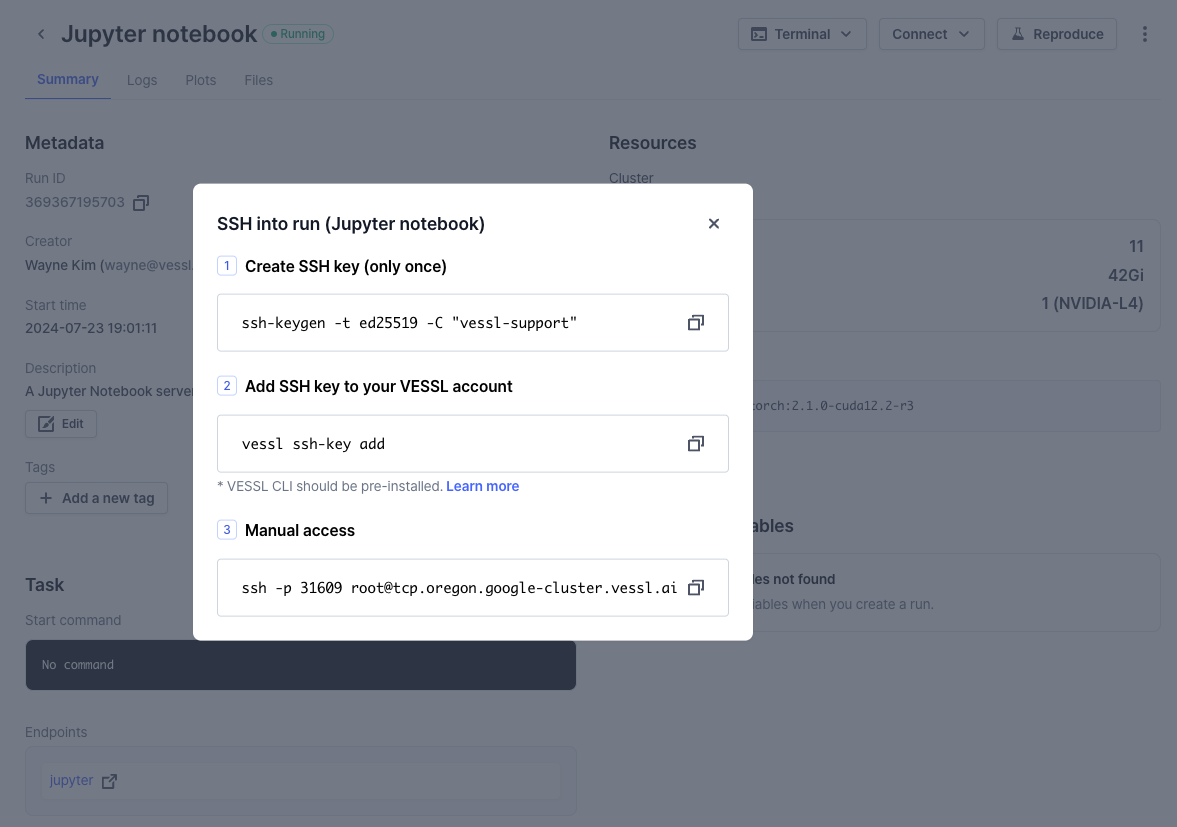

Manual access: The below command for manual access may change when you
perform a new run due to changes in IP address, user account, port number,
SSH key file path, proxy settings, or other options.
ssh -p 22 root@34.127.82.9Tips & tricks
Keep in mind that GPUs are fully dedicated to a notebook server —and therefore consume credits— even when you are not running compute-intensive cells. To optimize GPU usage, use tools like nbconvert to convert the notebook into a Python file or package it as a Python container and run it as a batch job. You can also mount volumes to interactive workloads by definingimport and reference files or datasets from your notebook.